Programming Assignment 2Date Posted: October 8
|
||||
| Background Information | ||||
|
In 1900 after almost forty years in obscurity the work of Gregor Mendel was
rediscovered by Hugo de Vries and Carl Correns. In 1902 Theodor Boveri, a
German geneticist, and Walter Sutton,
an American geneticist, independently established that genes are "carried" on chromosomes.
Each species of plant or animal has a certain number of chromosomes arranged in pairs in
each of their cells. One chromosome of each pair is
inherited from the father and one from the mother. Shortly after that two British geneticists, William Bateson and Reginald Punnett of St. Johnís College, Cambridge, discovered that certain traits always seemed to be inherited together. These traits, which seem in contradiction to the Mendelian Law of Independent Assortment, are called linked traits. They are almost always inherited together because they reside on the same chromosome. The key words here are "almost always inherited together". It was later discovered that through a process called crossing over certain segments of chromosomes can swap places, that is a segment of genes residing on one chromosome of a pair can swap places with the same segment on its' chromosome pair. It was also discovered that the frequency of such crossing over was related to the distance two genes were separated on the chromosome which they shared. Bateson, by the way, was the first to use the term genetics (from the Greek genno, 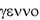 meaning to give birth) to describe the study of
heredity and biological inheritance. meaning to give birth) to describe the study of
heredity and biological inheritance. |
||||
| Your Assignment | ||||
|
You are to write a simulation program to
demonstrate the processes of inheritance incorporating the Mendelian laws
of genetics and later information on chromosomal structure and gene linking. The design and implementation of this program must follow object oriented design principles. Opportunity will be provided for you to question the customer's representative (the instructor) in class for more details. All requirements from programming assignment 1 are to be followed unless modified in the following list of requirements for this simulation.
|
||||
Structural Requirements
|
||||
| Deliverables | ||||
|
These products as specified below shall be delivered
electronically via e-mail to the instructor. Preliminary Class Diagram -- The class diagram shall be drawn using standard UML notation and shall show all of the classes to be implemented in the software and their relationships (dependencies, associations, generalizations, realizations, etc.) The PCD shall be submitted for instructor approval NLT (Not Later Than) Tuesday, October 22. Class Outline -- The class outline shall list all proposed variables and functions in each proposed class with a brief description of what each does. The class outline shall be submitted for instructor approval NLT (Not Later Than) Thursday, October 31. Functionality Outline -- The functionality outline shall be an outline which will show the step-by-step functionality of the program. This should be taken out to a fair amount of detail. The functionality outline shall be submitted for instructor approval NLT (Not Later Than) Thursday, November 7. Final Project -- The entire software project (compatible with Microsoft Visual Studio 2012 or 2015) shall be compressed into a zip file and submitted for instructor approval NLT Thursday, November 21. Just turning in your source files is not acceptable. |
||||
|
We will have several class periods in which we will meet as a team to discuss and plan this project. We will be doing a lot of brainstorming and planning together, but remember that each person is responsibility for implementing the final design on their own. |
||||
|
To download a sample executable as well as the sample data files and a parser for the data files click here. |
||||
| Input File Format | ||||
A data parser class (GeneticsSimDataParser.h and .cpp)
will be provided for reading, parsing, and providing data from
the data files. Two data files will be provided by the instructor
they will be in a modified XML format. Below is a sample of the
type of data file that will be used by the simulation.<!-- Sample data file for use in the Mendelian Genetics Simulation Program 2 --> <!-- Note: This file is not fully compatible with XML standards, but close enough --> <!-- for the purposes of this project --> <MENDELIAN_GENETICS_SIM> <ORGANISM> <GENUS> Pisum </GENUS> <SPECIES> Sativum </SPECIES> <COMMON_NAME> Pea Plant <COMMON_NAME> <CHROMOSOME_COUNT> 2 </CHROMOSOME_COUNT> </ORGANISM> <GENES> <GENE> <GENE_TRAIT> Plant Stature </GENE_TRAIT> <DOMINANT_ALLELE> Tall </DOMINANT_ALLELE> <DOMINANT_SYMBOL> T </DOMINANT_SYMBOL> <RECESSIVE_ALLELE> Dwarf </RECESSIVE_ALLELE> <RECESSIVE_SYMBOL> t </RECESSIVE_SYMBOL> <CROSSOVER_CHANCE> 5.2 </CROSSOVER_CHANCE> </GENE> <GENE> <GENE_TRAIT> Seed Texture </GENE_TRAIT> <DOMINANT_ALLELE> Wrinkled </DOMINANT_ALLELE> <DOMINANT_SYMBOL> W </DOMINANT_SYMBOL> <RECESSIVE_ALLELE> Smooth </RECESSIVE_ALLELE> <RECESSIVE_SYMBOL> w </RECESSIVE_SYMBOL> <CROSSOVER_CHANCE> 4.3 </CROSSOVER_CHANCE> </GENE> <GENE> <GENE_TRAIT> Seed Color </GENE_TRAIT> <DOMINANT_ALLELE> Green </DOMINANT_ALLELE> <DOMINANT_SYMBOL> S </DOMINANT_SYMBOL> <RECESSIVE_ALLELE> Yellow </RECESSIVE_ALLELE> <RECESSIVE_SYMBOL> s </RECESSIVE_SYMBOL> <CROSSOVER_CHANCE> 4.25 </CROSSOVER_CHANCE> </GENE> <GENE> <GENE_TRAIT> Flower Color </GENE_TRAIT> <DOMINANT_ALLELE> Purple </DOMINANT_ALLELE> <DOMINANT_SYMBOL> C </DOMINANT_SYMBOL> <RECESSIVE_ALLELE> White </RECESSIVE_ALLELE> <RECESSIVE_SYMBOL> c </RECESSIVE_SYMBOL> <CROSSOVER_CHANCE> 5.75 </CROSSOVER_CHANCE> </GENE> </GENES> <PARENTS> <PARENT> <CHROMOSOME> <STRAND1> T C </STRAND1> <STRAND2> t c </STRAND2> </CHROMOSOME> <CHROMOSOME> <STRAND1> W S </STRAND1> <STRAND2> w s </STRAND2> </CHROMOSOME> </PARENT> <PARENT> <CHROMOSOME> <STRAND1> T C </STRAND1> <STRAND2> t c </STRAND2> </CHROMOSOME> <CHROMOSOME> <STRAND1> W S </STRAND1> <STRAND2> w s </STRAND2> </CHROMOSOME> </PARENT> </PARENTS> <MENDELIAN_GENETICS_SIM> |
||||
| Output Format | ||||
The output printed on the screen shall contain the results of an experimental run in the following format:
Master Genes: Trait Name: Plant Stature Dominant Name: Tall(T) Recessive Name: Dwarf(t) Chance of crossover: 5.2 Trait Name: Seed Texture Dominant Name: Wrinkled(W) Recessive Name: Smooth(w) Chance of crossover: 4.3 Trait Name: Seed Color Dominant Name: Green(S) Recessive Name: Yellow(s) Chance of crossover: 4.25 Trait Name: Flower Color Dominant Name: Purple(C) Recessive Name: White(c) Chance of crossover: 5.75 Sim parent 1 Organism genus-species: Pisum Sativum Chromosomes: Chromosome 1 Gene Type: Plant Stature Allele 1: Tall(T) Allele 2: Dwarf(t) Gene Type: Flower Color Allele 1: Purple(C) Allele 2: White(c) Chromosome 2 Gene Type: Seed Texture Allele 1: Wrinkled(W) Allele 2: Smooth(w) Gene Type: Seed Color Allele 1: Green(S) Allele 2: Yellow(s) Sim parent 2 Organism genus-species: Pisum Sativum Chromosomes: Chromosome 1 Gene Type: Plant Stature Allele 1: Tall(T) Allele 2: Dwarf(t) Gene Type: Flower Color Allele 1: Purple(C) Allele 2: White(c) Chromosome 2 Gene Type: Seed Texture Allele 1: Wrinkled(W) Allele 2: Smooth(w) Gene Type: Seed Color Allele 1: Green(S) Allele 2: Yellow(s) How many offspring do you want to generate? (Type the number then press Enter) -->50 ======================= Results of this Run ======================= Gene: Plant Stature 11 homozygous dominant (Tall TT) 23 heterozygous dominant (Tall Tt) 16 homozygous recessive (Dwarf tt) Gene: Flower Color 8 homozygous dominant (Purple CC) 27 heterozygous dominant (Purple Cc) 15 homozygous recessive (White cc) Gene: Seed Texture 13 homozygous dominant (Wrinkled WW) 25 heterozygous dominant (Wrinkled Ww) 12 homozygous recessive (Smooth ww) Gene: Seed Color 12 homozygous dominant (Green SS) 25 heterozygous dominant (Green Ss) 13 homozygous recessive (Yellow ss) A total of 14 offspring had at least one crossover gene. |
||||